Evaluating submodels with the same model object
Source:vignettes/articles/Submodels.Rmd
Submodels.RmdSome R packages can create predictions from models that are different than the one that was fit. For example, if a boosted tree is fit with 10 iterations of boosting, the model can usually make predictions on submodels that have less than 10 trees (all other parameters being static). This is helpful for model tuning since you can cheaply evaluate tuning parameter combinations which often results in a large speed-up in the computations.
In parsnip, there is a method called multi_predict()
that can do this. It’s current methods are:
## [1] multi_predict._C5.0* multi_predict._earth*
## [3] multi_predict._elnet* multi_predict._glmnetfit*
## [5] multi_predict._lognet* multi_predict._multnet*
## [7] multi_predict._torch_mlp* multi_predict._train.kknn*
## [9] multi_predict._xgb.Booster* multi_predict.default*
## see '?methods' for accessing help and source codeWe’ll use the attrition data in rsample to
illustrate:
## ── Attaching packages ──────────────────────── tidymodels 1.4.1.9000 ──## ✔ broom 1.0.12 ✔ rsample 1.3.2
## ✔ dials 1.4.2 ✔ tailor 0.1.0
## ✔ dplyr 1.2.0 ✔ tidyr 1.3.2
## ✔ infer 1.1.0 ✔ tune 2.0.1
## ✔ modeldata 1.5.1 ✔ workflows 1.3.0
## ✔ purrr 1.2.1 ✔ workflowsets 1.1.1
## ✔ recipes 1.3.1 ✔ yardstick 1.3.2## ── Conflicts ──────────────────────────────── tidymodels_conflicts() ──
## ✖ purrr::discard() masks scales::discard()
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ✖ recipes::step() masks stats::step()
data(attrition, package = "modeldata")
set.seed(4595)
data_split <- initial_split(attrition, strata = "Attrition")
attrition_train <- training(data_split)
attrition_test <- testing(data_split)A boosted classification tree is one of the most low-maintenance approaches that we could take to these data:
# requires the xgboost package
attrition_boost <-
boost_tree(mode = "classification", trees = 100) |>
set_engine("C5.0")Suppose that 10-fold cross-validation was being used to tune the model over the number of trees:
set.seed(616)
folds <- vfold_cv(attrition_train)The process would fit a model on 90% of the data and predict on the
remaining 10%. Using rsample:
model_data <- analysis(folds$splits[[1]])
pred_data <- assessment(folds$splits[[1]])
fold_1_model <-
attrition_boost |>
fit_xy(x = model_data |> dplyr::select(-Attrition), y = model_data$Attrition)For multi_predict(), the same semantics of
predict() are used but, for this model, there is an extra
argument called trees. Candidate submodel values can be
passed in with trees:
fold_1_pred <-
multi_predict(
fold_1_model,
new_data = pred_data |> dplyr::select(-Attrition),
trees = 1:100,
type = "prob"
)
fold_1_pred## # A tibble: 111 × 1
## .pred
## <list>
## 1 <tibble [100 × 3]>
## 2 <tibble [100 × 3]>
## 3 <tibble [100 × 3]>
## 4 <tibble [100 × 3]>
## 5 <tibble [100 × 3]>
## 6 <tibble [100 × 3]>
## 7 <tibble [100 × 3]>
## 8 <tibble [100 × 3]>
## 9 <tibble [100 × 3]>
## 10 <tibble [100 × 3]>
## # ℹ 101 more rowsThe results is a tibble that has as many rows as the data being
predicted (n = 111). The .pred column contains a
list of tibbles and each has the predictions across the different number
of trees:
fold_1_pred$.pred[[1]]## # A tibble: 100 × 3
## trees .pred_No .pred_Yes
## <int> <dbl> <dbl>
## 1 1 0.916 0.0840
## 2 2 0.558 0.442
## 3 3 0.706 0.294
## 4 4 0.542 0.458
## 5 5 0.630 0.370
## 6 6 0.693 0.307
## 7 7 0.741 0.259
## 8 8 0.662 0.338
## 9 9 0.596 0.404
## 10 10 0.638 0.362
## # ℹ 90 more rowsTo get this into a format that is more usable, we can use
tidyr::unnest() but we first add row numbers so that we can
track the predictions by test sample as well as the actual classes:
fold_1_df <-
fold_1_pred |>
bind_cols(pred_data |> dplyr::select(Attrition)) |>
add_rowindex() |>
tidyr::unnest(.pred)
fold_1_df## # A tibble: 11,100 × 5
## trees .pred_No .pred_Yes Attrition .row
## <int> <dbl> <dbl> <fct> <int>
## 1 1 0.916 0.0840 No 1
## 2 2 0.558 0.442 No 1
## 3 3 0.706 0.294 No 1
## 4 4 0.542 0.458 No 1
## 5 5 0.630 0.370 No 1
## 6 6 0.693 0.307 No 1
## 7 7 0.741 0.259 No 1
## 8 8 0.662 0.338 No 1
## 9 9 0.596 0.404 No 1
## 10 10 0.638 0.362 No 1
## # ℹ 11,090 more rowsFor two samples, what do these look like over trees?
fold_1_df |>
dplyr::filter(.row %in% c(1, 88)) |>
ggplot(aes(x = trees, y = .pred_No, col = Attrition, group = .row)) +
geom_step() +
ylim(0:1) +
theme(legend.position = "top")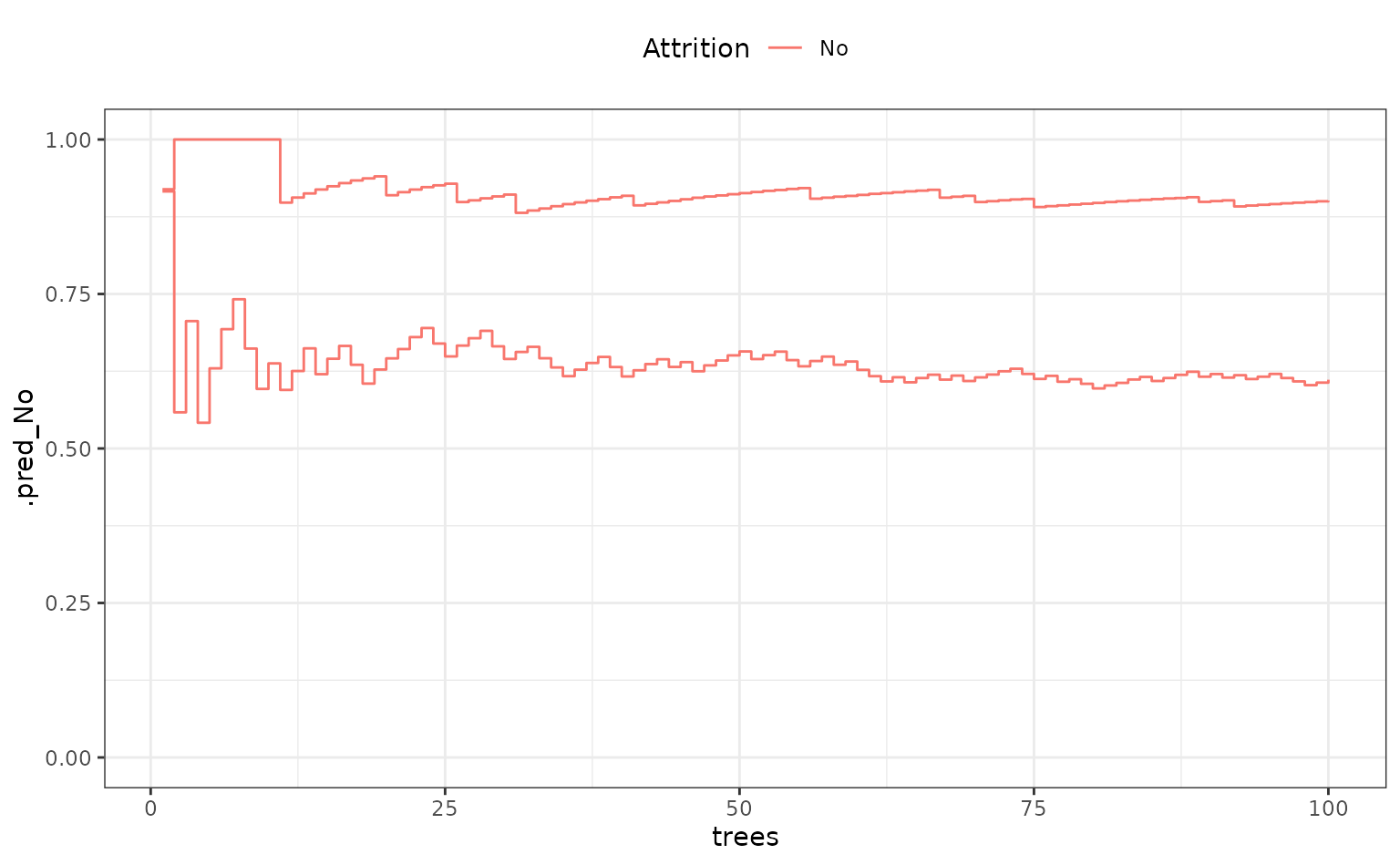
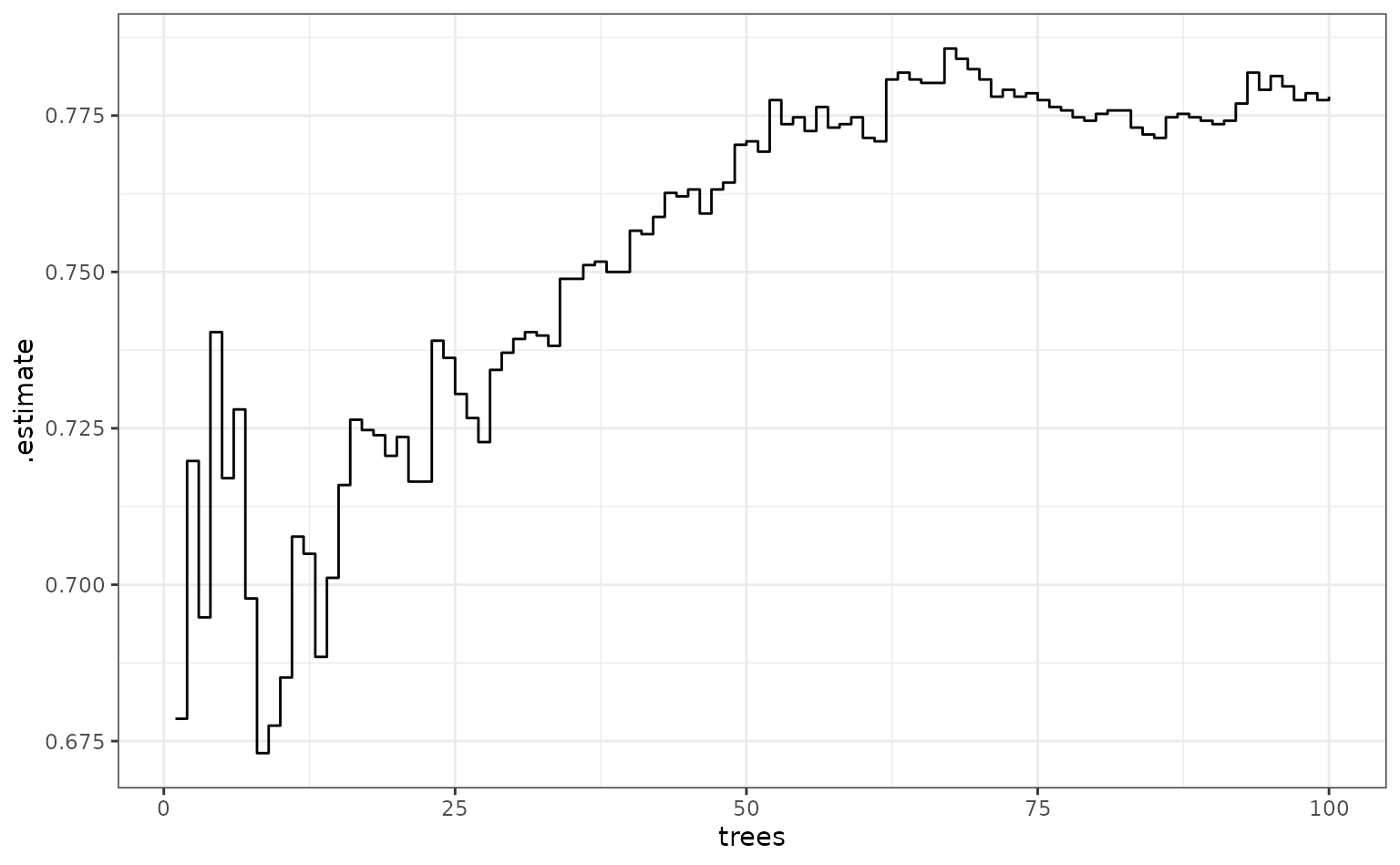
What does performance look like over trees (using the area under the ROC curve)?
fold_1_df |>
group_by(trees) |>
roc_auc(truth = Attrition, .pred_No) |>
ggplot(aes(x = trees, y = .estimate)) +
geom_step()